Retroviral uncoating is an area of great scientific interest. The recent finding that an intact HIV-1 mature core can by passing through nuclear pores, enter the nucleus of the infected cells, has shed new light on this process. By combining cryoEM, MS and iCLIP analysis, we investigate several aspects of the retroviral uncoating process, including the mature core organization and structure, nuclear pore transport and the involvement of host cofactors.
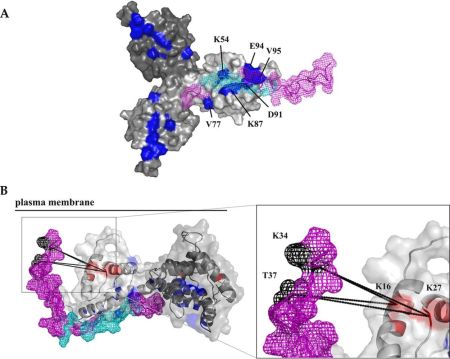
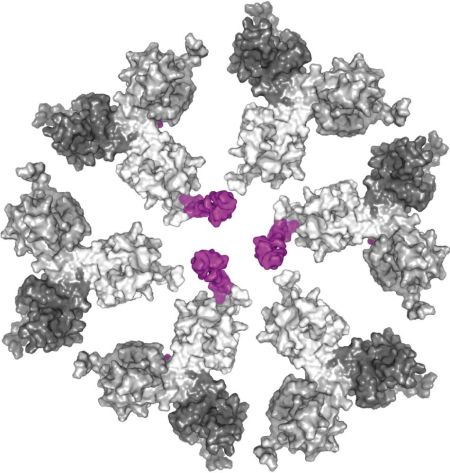
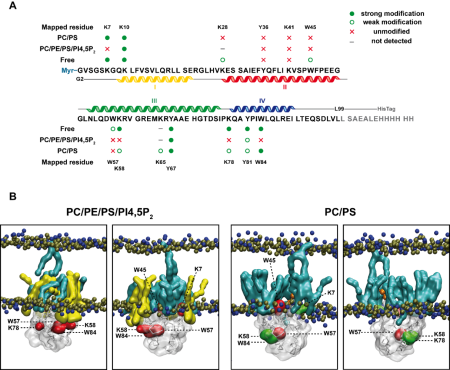
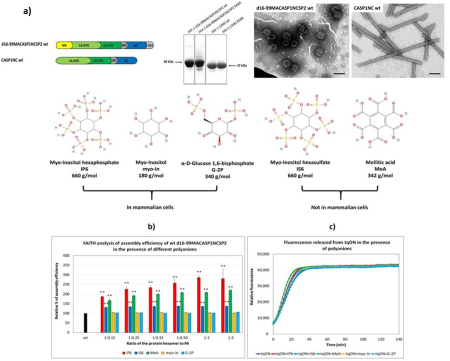
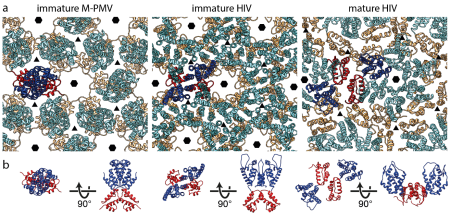
We are also interested in the step maturation or processing of the retroviral structural polyprotein Gag which is critical for the correct formation of a mature, fully infectious viral particle. Firstly, we focus on the last step of HIV-1 maturation, i.e. the cleavage of a short peptide (SP1) from the C-terminus of the capsid protein (CA). This step is inhibited by maturation inhibitors, new derivatives of which we have been testing by a combination of cellular and in vitro methods. Another area of our interest is the first step of Mason-Pfizer monkey virus (M-PMV) maturation, the cleavage of the N-terminal matrix domain (MA) from downstream located phosphoprotein (PP). This cleavage is thought to trigger structural changes accompanied by reorganization of the virus particle. Our new structural data show that interactions of the matrix domain (MA) of M-PMV with the plasma membrane lead to the exposure of
co-translationally attached myristoyl, and this conformational change subsequently significantly affects the initial maturation step. Therefore, these conformational changes may represent a control element in the sequential cleavage of the Gag polyprotein.